In Mac OS X, connection can directly be made to a remote FTP (File Transfer Protocol) server, from within the OS, without using any additional software.

Download files from Microsoft OneDrive to Mac via FTP. Browse to the destination folder on your Mac from the Local site box; select the files you want to download from your OneDrive account in the Remote site box, right click on them, and choose Download from the pop-up menu to download them from OneDrive to Mac via FTP. Firefox itself is able to download files from ftp sources. Here is my problem that I posted a couple days ago. 'John99 #answer-978193 said' Down grading Firefox is generally a bad idea, and you would be safer using an alternative browser.
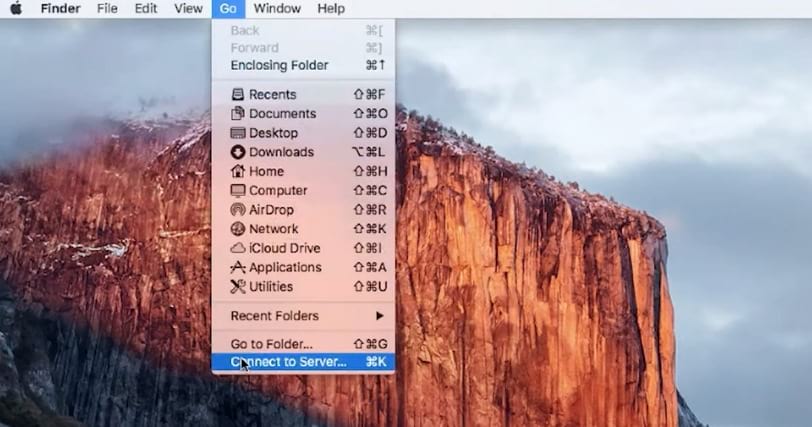
The easiest way is to open the Finder Window and select the from the main menu Go -> connect to Server. A dialog box will appear. Enter the server name (including ftp://) and click connect.
You can then login either as a guest user or enter username/password for registered accounts.

Alternatively, you can type the ftp server name in the Safari’s address bar for quick access to the built-in FTP client.
The downside of this simple method is that it can only be used for downloading files. Moreover, if the user name/password consists of the symbol ‘@‘ then, well, Finder fails to connect!
Briefly, I have FASTQ file of around 1 gigabyte and I want to convert it into FASTA format to be able to treat this data using MEGA 6 software. Therefore, I kindly want to ask how can I convert. Click “Select FASTQ File” and select FASTQ file in Open file dialog. Click “Select FASTA File” and select FASTA file in Save file dialog. Click “Convert” button to start conversion. Click “Cancel” to cancel conversion.
What are the highlights of the genomes FTP site?
The genomes FTP site offers a consistent core set of files for the genome sequence and annotation products of all organisms and assemblies in scope. It supports download needs such as:
- Retrieve the unmasked or soft-masked genome sequence for a specific genome assembly
- Retrieve GenBank or RefSeq Gene, RNA and protein annotation for a specific organism and a specific assembly, or a specific RefSeq annotation release
- Retrieve annotation in GenBank flat-file, GFF or GTF format
- Matching sequence identifiers in FASTA & GFF or GTF files to facilitate RNA-Seq and other analyses
- Confirm downloaded content is complete using provided md5checksums
What is the easiest way to download data for multiple genome assemblies?
The genome download service in the Assembly resource makes it easy to download data for multiple genomes without having to write scripts. To use the download service, run a search in Assembly, use facets to refine the set of genome assemblies of interest, open the 'Download Assemblies' menu, choose the source database (GenBank or RefSeq), choose the file type, then click the Download button to start the download. An archive file will be saved to your computer that can be expanded into a folder containing the genome data files from your selections.
For example, to download genomic FASTA sequence for all RefSeq bacterial complete genome assemblies:
- Start with an 'all[filter]' query on Assembly
- Select 'Bacteria' from the 'Organism group' facet in the left-hand sidebar
- Select 'Complete genome' from the 'Assembly level' facet in the left-hand sidebar
- Click on the 'Download Assemblies' button to open the download menu
- Leave 'Source database' set to RefSeq
- Select 'Genomic FASTA' from the 'File type' menu
- Wait for the 'calculating size.' message to be replaced by an estimated size
- Click Download, you may get a pop-up window asking if/where you want to save the genome_assemblies.tar archive file
- After the download has finished, expand the tar archive
- The resulting folder named 'genome_assemblies' will contain:
- a report.txt file that provides a summary of what was downloaded
- a folder named like 'ncbi-genomes-YYYY-MM-DD', where YYYY-MM-DD is the date of the download, containing:
- a README.txt file
- an md5checksums.txt file
- many data files with names like *_genomic.fna.gz, in which the first part of the name is the assembly accession followed by the assembly name
Simple variations on these steps can be used to obtain different file types or data for different sets of genome assemblies. If 'All file types (including assembly structure directory)' is selected from the 'File type' menu, the 'ncbi-genomes-YYYY-MM-DD' folder will contain a folder for each of the selected genome assemblies containing all the content from the FTP directory for that assembly.
The genome download service is best for small to moderately sized data sets. Selecting very large numbers of genome assemblies may result in a download that takes a very long time (depending on the speed of your internet connection). Scripting using rsync is the recommended protocol to use for downloading very large data sets (see below).
What is the best protocol to use to download large data sets?
We recommend using the rsync file transfer program from a Unix command line to download large data files because it is much more efficient than older protocols. The next best options for downloading multiple files are to use the HTTPS protocol, or the even older FTP protocol, using a command line tool such as wget or curl. Web browsers are very convenient options for downloading single files even though they will use the FTP protocol because of how our URLs are constructed. Other FTP clients are also widely available but do not all correctly handle the symbolic links used widely on the genomes FTP site (see below).
To use rsync
Replace the 'ftp:' at the beginning of the FTP path with 'rsync:'. E.g. If the FTP path is
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1, then the directory and its contents could be downloaded using the following rsync command:rsync --copy-links --recursive --times --verbose rsync://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1 my_dir/A file with FTP path
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gzcould be downloaded using the following rsync command:rsync --copy-links --times --verbose rsync://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gz my_dir/Diner dash 2.To use HTTPS
Replace the 'ftp:' at the beginning of the FTP path with 'https:'. Also append a '/' to the path if it is a directory. E.g. If the FTP path is
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1, then the directory and its contents could be downloaded using the following wget command:wget --recursive -e robots=off --reject 'index.html' --no-host-directories --cut-dirs=6 https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/ -P my_dir/A file with FTP path
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gzcould be downloaded using either of the following commands:wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gz -P my_dir/curl --remote-name --remote-time https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gzTo use FTP
Append a '/' to the path if it is a directory. E.g. If the FTP path is
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1, then the directory and its contents could be downloaded using the following wget command:wget --recursive --no-host-directories --cut-dirs=6 ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/ -P my_dir/A file with FTP path
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gzcould be downloaded using either of the following commands:wget --timestamping ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gz -P my_dir/curl --remote-name --remote-time ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/696/305/GCF_001696305.1_UCN72.1/GCF_001696305.1_UCN72.1_genomic.gbff.gzWhy was the NCBI genomes FTP site reorganized?
Historically, the genomes FTP site had been populated by different process flows and NCBI working groups leading to undesirable differences in available content and file formats. Also, data for GenBank genomes and RefSeq genomes were located in different areas of the NCBI FTP site that had different organization.
NCBI redesigned the genomes FTP site to expand the content and facilitate data access through an organized predictable directory hierarchy with consistent file names and formats. The site now provides greater support for downloading assembled genome sequences and/or corresponding annotation data with more uniformity across species. The current FTP site structure provides a single entry point to access content representing either GenBank or RefSeq data.
The initial release of the redesigned genomes FTP site in August 2014 added three new directories, namely ‘genbank’, ‘refseq’, and ‘all’ to the existing ftp area – ftp://ftp.ncbi.nlm.nih.gov/genomes/. These directories provide a core set of files representing both sequence and annotation content in several formats (see below). Additional file formats were added in subsequent updates.
The content of most of the old directories on the ftp://ftp.ncbi.nlm.nih.gov/genomes/ site, and the content previously at ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/ is no longer being updated. Many old directories from these two areas were moved to archival subdirectories within the /genomes/ area on 2 December 2015. Most of the remaining old directories were moved to the archive in March 2020. Details of what FTP directories and files were moved are as follows.
- All directories and files from ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/ were archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank
- The following directories from ftp://ftp.ncbi.nlm.nih.gov/genomes/ were archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_refseq/
- all Genus_species directories
- ASSEMBLY_BACTERIA
- Bacteria
- Bacteria_DRAFT
- Chloroplasts
- CLUSTERS
- Fungi
- MITOCHONDRIA
- PLANTS
- Plasmids
- Protozoa
- The file old_genomeID2nucGI from ftp://ftp.ncbi.nlm.nih.gov/genomes/ was archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/
- The IDS directory from ftp://ftp.ncbi.nlm.nih.gov/genomes/ was moved to ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/
How can I stay informed about changes to the NCBI genomes FTP site?
Subscribe to the genomes-announce mail list.
Are all genomes available in NCBI nucleotide available on the FTP site?
Genome sequence and annotation data is provided for organisms in scope for NCBI’s Assembly resource. Data are provided for both GenBank and RefSeq assembly versions. The FTP directories for the latest version in each assembly chain, and directories for many older assembly versions, include a core set of files and formats plus additional files relevant to the data content of the specific assembly. Directories for old assembly versions that predate the genomes FTP site reorganization contain only the assembly report, assembly stats & assembly status files.
Are files on the FTP site updated following annotation updates?
Yes, the FTP files for the latest version of an assembly are updated after the annotation on any of the sequences in the assembly changes.
The FTP files for the latest version of an assembly may also be updated:
- to make the files conform to the latest specifications for a particular data format
- to correct errors in conversion of the primary data from the NCBI databases into the various FTP file formats
Files for old versions of assemblies will not usually be updated, consequently, most users will want to download data only for the latest version of each assembly. For more information, see 'How can I download only the current version of each assembly?'.
My organism of interest is available in both GenBank and RefSeq. Is the genome the same? Which one should I use?
GenBank content includes genome assemblies that are submitted to members of the International Nucleotide Sequence Database Collaboration. GenBank submissions may or may not include annotation information which, when provided, was generated by different groups using different methods. Note that for prokaryotes, GenBank annotation may have been generated using NCBI’s prokaryotic genome annotation service. In contrast, RefSeq genomes are selected from, and are a subset of, the available GenBank genomes and annotation data is available for all RefSeq genomes, except for some viruses. RefSeq annotation content originates from NCBI’s prokaryotic, eukaryotic, organelle, or viral annotation pipelines, or is propagated from the GenBank submission.
For some assemblies, both GenBank and RefSeq content may be available. RefSeq genomes are a copy of the submitted GenBank assembly. In some cases the assemblies are not completely identical as RefSeq has chosen to add a non-nuclear organelle unit to the assembly or to drop very small contigs or reported contaminants. Equivalent RefSeq and GenBank assemblies, whether or not they are identical, and RefSeq to GenBank sequence ID mapping, can be found in the assembly report files available on the FTP site or by download from the Assembly resource.
How are the FTP directories structured?
The base structure of the genomes ftp site includes several main directory areas that provide sequence and annotation content, or report files. Sequence and annotation content is further organized by major taxonomic groupings, then by species, then by assembly. Sequence content is defined by the Assembly resource. The genomes FTP site provides directories for:
- GenBank content organized by taxonomic group, then by species and assembly
- RefSeq content organized by taxonomic group, then by species and assembly
- all (union of GenBank and RefSeq) organized by individual assembly
- Assembly reports
- Genome reports
Within the GenBank and RefSeq directories, the directory hierarchy is:
- Taxonomic group
- Genus_species
- All assemblies
- Individual assemblies
- Latest assembly versions
- Individual assemblies
- RefSeq representative genomes (if any)
- Individual assemblies
- RefSeq reference genomes (if any)
- Individual assemblies
- Annotation releases (for organisms annotated by the NCBI Eukaryotic Genome Annotation Pipeline)
- Data sets for each annotation release
- All assemblies
- Genus_species
The first layer of organization consists of the following directories:
- genbank: content includes primary submissions of assembled genome sequence and associated annotation data, if any, as exchanged among members of the International Nucleotide Sequence Database Collaboration, of which NCBI’s GenBank database is a member. The GenBank directory area includes genome sequence data for a larger number of organisms than the RefSeq directory area; however, some assemblies are unannotated. The subdirectory structure includes:
- archaea
- bacteria
- fungi
- invertebrate
- metagenomes
- other – this directory is only provided for GenBank and includes submissions of synthetic genomes.
- plant
- protozoa
- vertebrate_mammalian
- vertebrate_other
- viral
- refseq: content includes assembled genome sequence and RefSeq annotation data. All RefSeq genomes have annotation. RefSeq annotation data may be calculated by NCBI annotation pipelines or propagated from the GenBank submission. The RefSeq directory area includes fewer organisms than the GenBank directory area because not all genome assemblies are selected for the RefSeq project. Subdirectories include:
- archaea
- bacteria
- fungi
- invertebrate
- plant
- protozoa
- vertebrate_mammalian
- vertebrate_other
- viral
- mitochondrion [Content is from the RefSeq release FTP site.]
- plasmid [Content is from the RefSeq release FTP site.]
- plastid [Content is from the RefSeq release FTP site.]
- all: content is the union of GenBank and RefSeq assemblies. Two directories under 'all' are named for the accession prefix (GCA or GCF) and these directories contain another three levels of directories named for digits 1-3, 4-6 & 7-9 of the assembly accession. The next level is the data directories for individual assembly versions. 'all' contains many directories for old versions of assemblies; these are archival and will not be updated to add new file formats or to refresh the data.
A third directory, named 'annotation_releases' contains the products of the NCBI Eukaryotic Genome Annotation Pipeline. The data are organized first by taxonomy ID and then by annotation release ID. It is expected that many users will prefer to access the annotation release data using the paths under the 'refseq' directory that use the organism name. - ASSEMBLY_REPORTS: content consists of four summary report files that include meta-data details of all the latest GenBank assemblies, all the latest RefSeq assemblies, the historical GenBank assemblies, or the historical RefSeq assemblies. These summary files provide a ftp path that can be used to retrieve the sequence and annotation data. Another file provides the expected genome assembly size range for different species as applied to submissions to GenBank.
- GENOME_REPORTS: content consists of summary reports of genome sequencing projects, associated annotation statistics, and some defined reference datasets within the RefSeq project. Reports are provided by the Genomes resource.
- genomes
- genbank
- bacteria
- Bacillus_thuringiensis
- all_assembly_versions
- GCA_000008505.1_ASM850v1 – this directory layer is named using the pattern: [Assembly accession.version]_[assembly name]
- all_assembly_versions
- Bacillus_thuringiensis
- bacteria
- genbank
- genomes
- refseq
- vertebrate_mammalian
- Homo_sapiens
- all_assembly_versions
- latest_assembly_versions
- reference
- GCF_000001405.39_GRCh38.p13
- Homo_sapiens
- vertebrate_mammalian
- refseq
Example directory hierarchy:
The directory hierarchy for the Genbank Bacillus thuringiensis strain 97-27 genome, which has the assembly accession GCA_000008505.1 and default assembly name of ‘ASM850v1’ looks like this:
The directory hierarchy for the annotated human reference genome looks like this:
What is the file content within each specific assembly directory?
Assembly directories for all current assemblies, and for many previous assembly versions, include a core set of files and formats plus additional files relevant to the data content of the specific assembly. Vst nexus. Directories for old assembly versions that predate the genomes FTP site reorganization contain only the assembly report, assembly stats assembly status files. All data files are named according to the pattern:
[assembly accession.version]_[assembly name]_content.[format]The entries below have the format: filename, download menu name in parentheses, description.
assembly_status.txt
A text file reporting the current status of this version of the assembly ('latest', 'replaced', or 'suppressed'). Any assembly anomalies are also reported.
*_assembly_report.txt (Assembly structure report)
Tab-delimited text file reporting the name, role and sequence accession.version for objects in the assembly. The file header contains meta-data for the assembly including: assembly name, assembly accession.version, scientific name of the organism and its taxonomy ID, assembly submitter, and sequence release date.
*_assembly_stats.txt (Assembly statistics report)
Tab-delimited text file reporting statistics for the assembly including: total length, ungapped length, contig scaffold counts, contig-N50, scaffold-L50, scaffold-N50, scaffold-N75 scaffold-N90.
*_assembly_regions.txt (Assembly regions report)
Provided for assemblies that include alternate or patch assembly units. Tab-delimited text file reporting the location of genomic regions and listing the alt/patch scaffolds placed within those regions.
*_assembly_structure directory
Contains AGP files that define how component sequences are organized into scaffolds and/or chromosomes. Other files define how scaffolds and chromosomes are organized into non-nuclear and other assembly-units, and how any alternate or patch scaffolds are placed relative to the chromosomes. Only present if the assembly has internal structure.
*_cds_from_genomic.fna.gz (CDS from genomic FASTA)
FASTA format of the nucleotide sequences corresponding to all CDS features annotated on the assembly, based on the genome sequence.
*_feature_count.txt.gz (Feature count)
Tab-delimited text file reporting counts of gene, RNA, CDS, and similar features, based on data reported in the *_feature_table.txt.gz file.
*_feature_table.txt.gz (Feature table)
Tab-delimited text file reporting locations and attributes for a subset of annotated features. Included feature types are: gene, CDS, RNA (all types), operon, C/V/N/S_region, and V/D/J_segment. Replaces the .ptt .rnt format files that were provided in the old genomes FTP directories.
*_genomic.fna.gz (Genomic FASTA)
FASTA format of the genomic sequence(s) in the assembly. Repetitive sequences in eukaryotes are masked to lower-case. The genomic.fna.gz file includes all top-level sequences in the assembly (chromosomes, plasmids, organelles, unlocalized scaffolds, unplaced scaffolds, and any alternate loci or patch scaffolds). Scaffolds that are part of the chromosomes are not included because they are redundant with the chromosome sequences; sequences for these placed scaffolds are provided under the assembly_structure directory.
*_genomic.gbff.gz (Genomic GenBank format)
GenBank flat file format of the genomic sequence(s) in the assembly. This file includes both the genomic sequence and the CONTIG description (for CON records), hence, it replaces both the .gbk .gbs format files that were provided in the old genomes FTP directories.
*_genomic.gff.gz (Genomic GFF)
Annotation of the genomic sequence(s) in Generic Feature Format Version 3 (GFF3). Sequence identifiers are provided as accession.version. Additional information about NCBI's GFF files is available at ftp://ftp.ncbi.nlm.nih.gov/genomes/README_GFF3.txt.
*_genomic.gtf.gz (Genomic GTF)
Annotation of the genomic sequence(s) in Gene Transfer Format Version 2.2 (GTF2.2). Sequence identifiers are provided as accession.version.
*_genomic_gaps.txt.gz (Genomic gaps)
Tab-delimited text file reporting the coordinates of all gaps in the top-level genomic sequences. The gaps reported include gaps specified in the AGP files, gaps annotated on the component sequences, and any other run of 10 or more Ns in the sequences.
*_protein.faa.gz (Protein FASTA)
FASTA format of the accessioned protein products annotated on the genome assembly.
*_protein.gpff.gz (Protein GenPept format)
GenPept format of the accessioned protein products annotated on the genome assembly.
*_rm.out.gz (RepeatMasker output)
RepeatMasker output; Provided for eukaryotes.
*_rm.run (RepeatMasker run info)
Documentation of the RepeatMasker version, parameters, and library (text format); Provided for eukaryotes.
*_rna.fna.gz (RNA FASTA)
FASTA format of accessioned RNA products annotated on the genome assembly; Provided for RefSeq assemblies as relevant (Note, RNA and mRNA products are not instantiated as a separate accessioned record in GenBank and are provided for some RefSeq genomes, most notably the eukaryotes.).
*_rna.gbff.gz (RNA GenBank format)
GenBank flat file format of RNA products annotated on the genome assembly; Provided for RefSeq assemblies as relevant.
*_rna_from_genomic.fna.gz (RNA from genomic FASTA)
FASTA format of the nucleotide sequences corresponding to all RNA features annotated on the assembly, based on the genome sequence.
*_translated_cds.faa.gz (Translated CDS)
FASTA sequences of individual CDS features annotated on the genomic records, conceptually translated into protein sequence. The sequence corresponds to the translation of the nucleotide sequence provided in the *_cds_from_genomic.fna.gz file.
*_wgsmaster.gbff.gz (WGS-master)
GenBank flat file format of the WGS master for the assembly (present only if a WGS master record exists for the sequences in the assembly).
annotation_hashes.txt
Tab-delimited text file reporting hash values for different aspects of the annotation data. The hashes are useful to monitor for when annotation has changed in a way that is significant for a particular use case and warrants downloading the updated records.
md5checksums.txt
File checksums are provided for all data files in the directory.
What additional files are provided for RefSeq genomes annotatated by the NCBI Eukaryotic Genome Annotation Pipeline?
Assembly directories for RefSeq genomes annotated by the NCBI Eukaryotic Genome Annotation Pipeline include extra sub-directories and files in additon to the standard set of files and formats. All data files are named according to the pattern:
[assembly accession.version]_[assembly name]_content.[format]The entries below have the format: filename, download menu name in parentheses, description.
Assembly directory
*_pseudo_without_product.fna.gz (Pseudo without product FASTA)
FASTA format of the genomic sequence corresponding to pseudogene and other gene regions which do not have any associated transcribed RNA products or translated protein products. It includes annotated gene regions that require rearrangement to provide the final product, e.g. immunoglobulin segments. These sequences are not assigned accession numbers, and are derived directly from the assembled genomic sequences. The FASTA title has a local sequence identifier, the Gene ID and gene name.
Evidence_alignments sub-directory
*_cross_species_tx_alns.gff.gz (Evidence alignments)
Alignments of cDNAs, ESTs and TSAs from other species to the genomic sequence(s) in Generic Feature Format Version 3 (GFF3) [not all annotation releases have cross-species alignments]. These alignments may have been used as evidence for gene prediction by the annotation pipeline. Sequence identifiers are provided as accession.version. Additional information about NCBI's GFF files is available at ftp://ftp.ncbi.nlm.nih.gov/genomes/README_GFF3.txt.
*_same_species_tx_alns.gff.gz (Evidence alignments)
Alignments of same-species cDNAs, ESTs and TSAs to the genomic sequence(s) in Generic Feature Format Version 3 (GFF3). These alignments were used as evidence for gene prediction by the annotation pipeline. Sequence identifiers are provided as accession.version. Additional information about NCBI's GFF files is available at ftp://ftp.ncbi.nlm.nih.gov/genomes/README_GFF3.txt.
Gnomon_models sub-directory
*_gnomon_model.gff.gz (Gnomon model GFF)
Gnomon annotation of the genomic sequence(s) in Generic Feature Format Version 3 (GFF3). Sequence identifiers are provided as accession.version for the genomic sequences and Gnomon identifiers for the Gnomon models: gene.XXX for genes, GNOMON.XXX.m for transcripts and GNOMON.XXX.p for proteins. These identifiers are NOT universally unique. They are unique per annotation release only. Additional information about NCBI's GFF files is available at ftp://ftp.ncbi.nlm.nih.gov/genomes/README_GFF3.txt.
*_gnomon_protein.faa.gz (Gnomon model protein FASTA)
FASTA format sequences of Gnomon protein models annotated on the genome assembly. The FASTA title is the Gnomon identifier for the protein model (>gnl GNOMON XXX.p).
*_gnomon_rna.fna.gz (Gnomon model RNA FASTA)
FASTA format sequences of Gnomon transcript models annotated on the genome assembly. The FASTA title is the Gnomon identifier for the transcript (>gnl GNOMON XXX.m).
RefSeq_transcripts_alignments sub-directory
*_knownrefseq_alns.bam (RefSeq transcript alignments)
Alignments of the annotated Known RefSeq transcripts (identified with accessions prefixed with NM_ and NR_) to the genome in BAM format [not all annotation releases have Known RefSeq transcripts]. For more information about the BAM format see: https://samtools.github.io/hts-specs/SAMv1.pdf.
*_knownrefseq_alns.bam.bai (RefSeq transcript alignments)
Index of the BAM alignments of the annotated Known RefSeq transcripts to the genome. [not all annotation releases have Known RefSeq transcripts].
*_modelrefseq_alns.bam (RefSeq transcript alignments)
Alignments of the annotated Model RefSeq transcripts (identified with accessions prefixed with XM_ and XR_) to the genome in BAM format. For more information about the BAM format see: https://samtools.github.io/hts-specs/SAMv1.pdf.
*_modelrefseq_alns.bam.bai (RefSeq transcript alignments)
Index of the BAM alignments of the annotated Model RefSeq transcripts to the genome.
Annotation_comparison sub-directory
This directory is only provided for re-annotations of the same species.
*_compare_prev.txt.gz (Annotation comparison report)
Matching genes and transcripts in the current and previous annotation releases binned by type of difference (column 1 for genes and column 14 for transcripts), in tabular format.
*_compare_prev.gbp.gz (Annotation comparison GenomeWorkBench)
Genome Workbench project file for visualization and search of differences between the current and previous annotation releases. The NCBI Genome Workbench web site provides help on downloading and using the 64-bit version of Genome Workbench.
What is the content of annotation_releases in the refseq directory hierarchy?
The annotation_releases directory provides data for specific annotation releases (100, 101, etc.) for organisms that have been annotated by the NCBI Eukaryotic Genome Annotation Pipeline. Each annotation release corresponds to an annotation run. The annotation release identifiers (AR) are numbered sequentially starting at 100,independently of the assembly used. An assembly may have been annotated multiple times, and be featured in different annotation release directories. The 'current' directory contains the data for the most recent annotation. For many organisms, only the most recent annotation may be available. Previous annotations are available at ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_refseq/.
Each annotation release directory contains:
README_[organism_name]_annotation_release_[annotation_release_id]
This file provides information specific to the specific annotation release, including data freeze dates, release date and release number, and the annotated assemblies.
[organism name]_ARXXX_annotation_report.xml
This file is the XML version of the HTML report for the organism, e.g. www.ncbi.nlm.nih.gov/genome/annotation_euk/Homo_sapiens/108/. It contains information on the annotation release, including:
- Important dates associated with the annotation
- Assemblies
- Gene and feature statistics
- Masking results
- Transcript and protein alignments used for the annotation
- Assembly-assembly alignments used to track genes from the previous assembly to the current, or from the reference to an alternate assembly if relevant
Assembly directory
One directory for each genome assembly that was annotated in the release. Named as [assembly accession.version]_[assembly name]. This directory contains the files provided for all genome assemblies plus those additional files provided for organisms annotated by the NCBI Eukaryotic Genome Annotation Pipeline.
How can I find the sequence and annotation of my genome of interest?
Genome assemblies of interest can be found using one of two methods.
Using the NCBI Assembly resource
Genome assemblies of interest can be found using the search bar, advanced search page or browse by organism table provided by the Assembly resource
GenBank or RefSeq data for the assembly can be obtained by following the links to the FTP site from the 'Access the data' section of the right-hand sidebar.
Using the assembly summary report files
Download the relevant assembly summary files that report assembly meta-data.
Search the meta-data fields, or filter the files, to find assemblies of interest (see README_assembly_summary.txt for a description of the columns) .
The field named 'ftp_path' provides the path to the FTP directory containing the data for each assembly.
- Either the two master assembly summary files:
ftp://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/assembly_summary_genbank.txt
ftp://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/assembly_summary_refseq.txt - Or an assembly summary file for a taxonomic group from the appropriate directory under genbank or refseq. e.g.
ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/assembly_summary.txt - Or an assembly summary file for a species from the appropriate directory under genbank or refseq. e.g.
ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/Salmonella_enterica/assembly_summary.txt
- Either the two master assembly summary files:
Where can I find information to help me chose between the many different assemblies for a species?
There can be many different genome assemblies available for species with medical, agricultural or scientific relevance. The Genus_species directories under the 'genbank' and 'refseq' directory trees each contain an assembly_summary.txt file that provides general information on all assembly versions included in the directory, such as release date, submitter organization, assembly level and status. See for example ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/archaea/Sulfolobus_islandicus/assembly_summary.txt
After assemblies of interest have been identified using the data from the species-specific assembly_summary.txt file, they can be accessed via the 'all_assembly_versions' directory for that species.
Alternatively, any assemblies that the NCBI Reference Sequence (RefSeq) group has selected to be reference or representative genomes can be readily accessed via the directories named 'reference' or 'representative' in the Genus_species directories under the 'genbank' and 'refseq' directory trees.
How can I download only the current version of each assembly?
Any changes to the sequences included in a particular assembly accession result in an increment of the assembly version, which means that an assembly accession.version (e.g. GCF_000001405.28) represents a fixed set of sequences. It also means that a particular assembly may have several versions, where only the most recent version is considered to be 'latest', and earlier versions are marked as either 'replaced' or 'suppressed'. In some cases the last version of an assembly may be 'suppressed', for example if it was removed from the RefSeq collection due to changes in scope or quality concerns.
Only FTP files for the 'latest' version of an assembly are updated when annotation is updated, new file formats are added or improvements to existing formats are released. Consequently, most users will want to download data only for the latest version of each assembly. You can select data from only the latest assemblies in several ways:
- Use the Assembly database and select the 'Latest' filter from the left sidebar, or add the term 'AND 'latest'[Filter]' to your query.
- Use the /genbank or /refseq FTP paths to navigate to the species level directory and then select assemblies from the 'latest_assembly_versions' subdirectory. See 'How are the FTP directories structured?' for more details.
- Use either of the two master assembly summary files, or the assembly_summary.txt file for the species or taxonomic group of interest (see above), select those assemblies that are marked as 'latest' in the version_status column (11), and then use the FTP path indicated in column 20 to download the data.
How can I download RefSeq data for all complete bacterial genomes?
The easiest way to download RefSeq data for all complete bacterial genomes is the use the genome download service in the Assembly resource, as described above.
Alternatively, the assembly summary report files provide information that can be used to identify a set of assemblies of interest along with their FTP file paths. For example, to obtain the GenBank flat file format annotation for all complete bacterial genomes in the NCBI Reference Sequences collection (RefSeq):
Variants of these instructions can be used to download all draft bacterial genomes in RefSeq (assembly_level is not 'Complete Genome'), all RefSeq reference or representative bacterial genomes (refseq_category (column 5) is 'reference genome' or 'representative genome'), etc.
- Download the /refseq/bacteria/assembly_summary.txt file
- List the FTP path (column 20) for the assemblies of interest, in this case those that have 'Complete Genome' assembly_level (column 12) and 'latest' version_status (column 11). One way to do this would be using the following awk command:
awk -F 't' '$12'Complete Genome' $11'latest'{print $20}' assembly_summary.txt > ftpdirpaths - Append the filename of interest, in this case '*_genomic.gbff.gz' to the FTP directory names. One way to do this would be using the following awk command:
awk 'BEGIN{FS=OFS='/';filesuffix='genomic.gbff.gz'}{ftpdir=$0;asm=$10;file=asm'_'filesuffix;print ftpdir,file}' ftpdirpaths > ftpfilepaths - Use a script to download the data file for each FTP path in the list
How can I download all genome assemblies from the Human Microbiome Project, or other project?
All genomes assemblies linked to a particular BioProject can be downloaded using the genome download service in the Assembly resource described above.
The following example will download all reference genomes for the Human Microbiome Project (HMP), which has the BioProject accession PRJNA28331.
- Search in BioProject for PRJNA28331
- Follow the link to 'Assembly' under 'Related information' in the right-hand sidebar
- Click on the 'Download Assemblies' button to open the download menu
- Select the 'Source database', either GenBank or RefSeq
- Select a 'File type', e.g. 'Genomic FASTA'
- Wait for the 'calculating size.' message to be replaced by an estimated size
- Click Download, you may get a pop-up window asking if/where you want to save the genome_assemblies.tar archive file
- After the download has finished, expand the tar archive
Why was the sequence identifier format in the FASTA files changed?
We changed the sequence identifier format in the FASTA files to make our datasets more usable by the community.
NCBI has traditionally used a compound FASTA sequence identifier string in which multiple IDs were separated by ' ' characters. This format provides more information but requires that the individual sequence identifiers be parsed out of the compound string. The FASTA files on the redesigned genomes FTP site have a simple sequence identifier string that is just the sequence accession.version, for example:
>U00096.3 Escherichia coli str. K-12 substr. MG1655, complete genome
>NC_000001.11 Homo sapiens chromosome 1, GRCh38 Primary AssemblyThis sequence identifier is identical to that used in the GFF and GTF annotation files on the genomes FTP site. Providing sequence and annotation files with matching sequence identifiers supports their use in commonly used RNA-Seq analysis packages and in other analysis pipelines that rely on simple string comparison to match sequence identifiers.
Why do some species directory names start with an underscore?
Certain symbols and punctuation marks have a special meaning to computer operating systems, consequently, they can cause problems if they are included as part of directory or file names. Examples include spaces, (, ), [, ] and '. Whenever one or more of these special characters appears in the organism name they are replaced by an underscore.
Taxonomy places square brackets around the genus for some species to indicate that they are misclassified. The current names continue to be used with square brackets until the species has been formally renamed. The square brackets around the genus are converted to underscores when a directory name is created for one of these misclassified species resulting in a directory name that begins with an underscore.
Do you provide assembly data formatted for use by sequence read alignment pipelines?
Genomic FASTA with modified sequence identifiers and index files convenient for analysis with Next Generation Sequencing tools are currently provided for the Genome Reference Consortium's human and mouse assemblies: GRCh38, GRCm38.p3 and GRCm39. RefSeq annotation in GFF3 and GTF formats with sequence identifiers matching those in the FASTA files are also provided to facilitate use in RNA-Seq analysis pipelines.
The four analysis sets provided for GRCh38 (no_alt_analysis_set, full_analysis_set, full_plus_hs38d1_analysis_set, no_alt_plus_hs38d1_analysis_set) and the two analysis sets provided for GRCm38.p3 (no_alt_analysis_set, full_analysis_set) differ from the corresponding full assemblies by one or more of the following:
- omission of alternate locus and patch scaffolds that cause complications for sequence read alignment programs that are not alt-aware
- hard masking of duplicate copies the pseudo-autosomal regions and centromeric arrays
- addition of 'decoy' sequences
Index files generated by BWA, Samtools, Bowtie and HISAT2 are provided. See the GRCh38 README, GRCm38 README or GRCm39 README for a full description.
Are repetitive sequences in eukaryotic genomes masked?
Repetitive sequences in eukaryotic genome assembly sequence files, as identified by WindowMasker, have been masked to lower-case.
The location and identity of repeats found by RepeatMasker are also provided in a separate file. These spans could be used to mask the genomic sequences if desired. Be aware, however, that many less studied organisms do not have good repeat libraries available for RepeatMasker to use.
How do alignment programs treat the lower-case masking in genomic fasta files?
Alignment programs typically have parameters that control whether the program will ignore lower-case masking, treat it as soft-masking (i.e. only for finding initial matches) or treat it as hard-masking. The program's documentation should indicate the default behavior.
By default NCBI BLAST will ignore lower-case masking but this can be changed by adding options to the blastn command-line.
To have blastn treat lower-case masking in the query sequence as soft-masking add:To have blastn treat lower-case masking in the query sequence as hard-masking add:
How can sequence with lower-case masking be converted to unmasked sequence?
Here are two examples of commands that will remove lower-case masking:
-or-
How can sequence with lower-case masking be converted to sequence masked with Ns?
Here are two examples of commands that will convert lower-case masking to masking with Ns (hard-masked):
-or-
Firefox truncates long FTP directory and file names. How can I see the full names?
The Firefox web browser is unable to display long FTP directory and file names in http mode. The problem can be circumvented by changing the URL from 'http://ftp.' to 'ftp://ftp.'.
Do ftp://ftp.ncbi.nlm.nih.gov/ and ftp://ftp.ncbi.nih.gov/ provide the same content?
These two paths are equivalent hence they do currently provide the same content, however, ftp://ftp.ncbi.nlm.nih.gov/ is the preferred path and the abbreviated path, ftp://ftp.ncbi.nih.gov/, may not be supported indefinitely.
Why does my FTP client not handle some FTP directories or files correctly?
The NCBI genomes FTP site makes extensive use of symbolic links to provide alternative paths to the same FTP files without duplicating the data. Many FTP clients have incomplete implementation of the FTP symbolic link specification or other bugs causing them to incorrectly treat symbolic links as files or directories. This may lead to the following problems:
- a symbolic link to a file is presented as a folder/directory
- a symbolic link to a directory is presented as a file
- never-the-less, clicking on the 'file' may still reveal it to be a folder/directory that can be browsed
- a symbolic link is copied as an alias instead of being resolved
To avoid these problems: How to download sims 4 disc on mac.
- download files using either the rsync or HTTPS protocols instead of the FTP protocol (see above)
- if using wget, append a '/' after the directory/folder name
- try a different FTP client:
- use a web browser that correctly shows what is a file, a directory or a symlink, such as Chrome or Firefox
- for FileZilla
- Windows: use the latest version of FileZilla
- Mac OSX: the bug causing symlinks to be shown as files has been reported on FileZilla ticket #4490 but has not yet been fixed
How To Download A Fasta File On Mac File
How To Download A Fasta File
How To Ftp From A Mac
- Genome Compiler is an all-in-one free software platform for biologists. You can intuitively visualize & design DNA, import, manage and share your data.
- Navigate to the location where you want the files to be stored. Click the Select button. Click the Close button to exit Preferences. To download a specific image that appears on a web page, move your pointer over the image, right-click, and choose Save Image As from the pop-up menu that appears.